Science
1.) Research objectives
Microbiomas are strongly associated with our health; However, the knowledge is still very limited.
Recently developed high-throughput sequencing techniques allow us to explore previously unexplored habitats for their microorganisms.
We selected apples because their microbiome has not been internationally studied, but it can have a strong impact on both human and plant health.
The project will look for green solutions for apple diseases and storage problems. The research goals are achieved through active participation of students. This provides an important opportunity to combine current research from the internationally recognized standard with pre-university promotion of junior staff. Students not only learn science, but also access to private garden or grocery items, allowing broader high throughput screening of plant disease antagonists.
The composition of apple-associated microbiomes derived from organic and conventional crops will be compared using microbiological, molecular and microscopic methods. In addition, differences in apple microbiomes of disease-degraded and healthy apple trees as well as of apples stored differently for the first time should be recorded.
An important goal of the project is the communication of science and its implementation in our society. This will be achieved sustainably by accompanying art projects, bringing research topics closer to students and interested people. This innovative interdisciplinary approach would not be feasible without the support of the Sparkling Science Program.
2.) State of knowledge
A. For the discovery of microbiomas.
While single microorganisms (MO) could be visualized by Antoni van Leeuwenhoek after the invention of the microscope in the 17th century, the identity of individual microorganisms and the structure of complex microbial communities remained unknown for a long time.
It was only at the end of the 20th century that the technique was developed that allows molecular fingerprints of microorganism communities to be mapped (Muyzer & Smalla 1998). This technique has already produced a first flowering of microbial ecology, as it has now enabled more (approximately 20%) of existing microorganisms to be detected. With hitherto customary cultivation methods, a maximum of 3% of them were detectable.
Only high-throughput sequencing of DNA made it possible to capture the full extent of microbial diversity.
Studies conducted in the past decade have revealed a high number of surprising diversity in microbial ecosystems. For example, more than 100 trillion microorganisms live in the human gut; a number ten times larger than the human body has cells (Qin et al., 2010). The number of species of microorganisms colonizing the human body is estimated at more than 10,000.
Similar dimensions are reported for plants: in the plant root, which, like our gut, serves to absorb nutrients, more microorganisms live than humans on earth (Philippot et al., 2013).
The complex microorganism communities are referred to as microbiome. In addition to this impressive diversity, the important functions of microorganisms contribute to a new perspective (Lozupone et al., 2012).
The image change of microorganisms can be described as a paradigm shift: all eukaryotes define themselves as meta-organisms and should only be considered in the context of their microbiome (Jones 2013). It also includes the discovery that pathogens represent only a tiny fraction of microorganisms; Often only the balance is shifted here. Scientists are therefore calling for a rethinking of microorganisms (Blaser 2011). This rethinking is also important to the population and should be knowledge-based.
Initial findings revealed a large and often unexpected diversity in microbiomes, although the influencing factors are often unknown. Especially in the context of food, application of micro-organisms for crop protection and stress prevention is currently a agricultural revolution.
Classic monitoring in the food sector allows only a small proportion of the diversity to be evaluated and thus does not provide any representative statements. The research into healthy foods with fewer pesticides has great potential for the development of healthy food.
It is of particular importance for us to explore these relationships and to make them understandable and tangible for the young people and.
B. The apple as a research object for our health.
Co-involvement of the human microbiome has been identified in many diseases, including cardiovascular disease, allergies and cancer (Qin et al., 2010).
Plant health is now also associated with the associated microorganisms: without them, many are unable to germinate, grow or produce tasty fruits (Berg et al., 2013, 2016).
Interestingly, there is a connection between the two microbiomes: not only does our diet determine our own individual microbiome directly through its composition, it also serves as an important source of beneficial microorganisms (Thomas et al., 2011). Although this fact is important for a healthy gut microbiome and can help prevent diseases such as inflammation and obesity (Le Chatelier et al., 2013), the microbiome on our food is unknown, with a few exceptions (Leff and Fierer 2013).
The National Science and Technology Council’s (November 2015) latest report named food microbiome research as the number one research priority in international microbiome research. In western countries, in addition to grapes and oranges, apples are an important source of dietary intake of fruits (Perez et al., 2001). Epidemiological studies indicate an inverse correlation between regular apple consumption and the risk of different cancers, cardiovascular disease and type II diabetes (Jedrychowski et al., 2009).
The health-promoting properties of apples as well as other plant foods that are consumed raw could be due, among other things, to the associated microbiome. The microbiota of the apple tree rhizosphere (directly affected by a living root in the Soil) as well as apple blossoms have been extensively researched by several research groups in recent years (Shade et al 2013, Caputo et al 2015). However, we are not aware of any study that deals with the apple fruit microbiome, which underlines the innovation and relevance of our project.
One of the most common diseases of apple trees in Europe is the apple scab (MacHardy 1996, Fig. 1), which is caused by the fungus Venturia inaequalis and results in yield losses of 70% and can lead higher.
Gloeosporium pillars on the apple is an important storage disease that can be caused by three different fungi. In recent years, the Venturia resistance to an increasing number of chemical fungicides has grown in large apple-growing areas.
An environmentally friendly alternative to chemical pesticides is biocontrol, a process of using living organisms to control pests (Berg 2009).
The biological control of apple scab has been researched for 50 years (Carisse 2000). A new study has shown that a fungus ( Cladosporium cladosporioides H39) is capable of sporulating V. inaequalis to reduce significantly. An effect of H39 in field trials in commercial orchards could even be demonstrated (Köhl et al., 2015). Nevertheless, there are still no microbiological products against this disease on the market (Carisse 2000, Köhl et al., 2015).
3.) Scientific
Approach
With the help of microbiological and molecular biological or microscopic analyzes we want to search for the diversity of microbiological communities on and in biologically and conventionally grown, healthy and disease-injured apples.
Many of the selected methods were used in the workgroup of Univ. Prof. Gabriele Berg, who has internationally recognized expertise in the field of plant microbiome research.
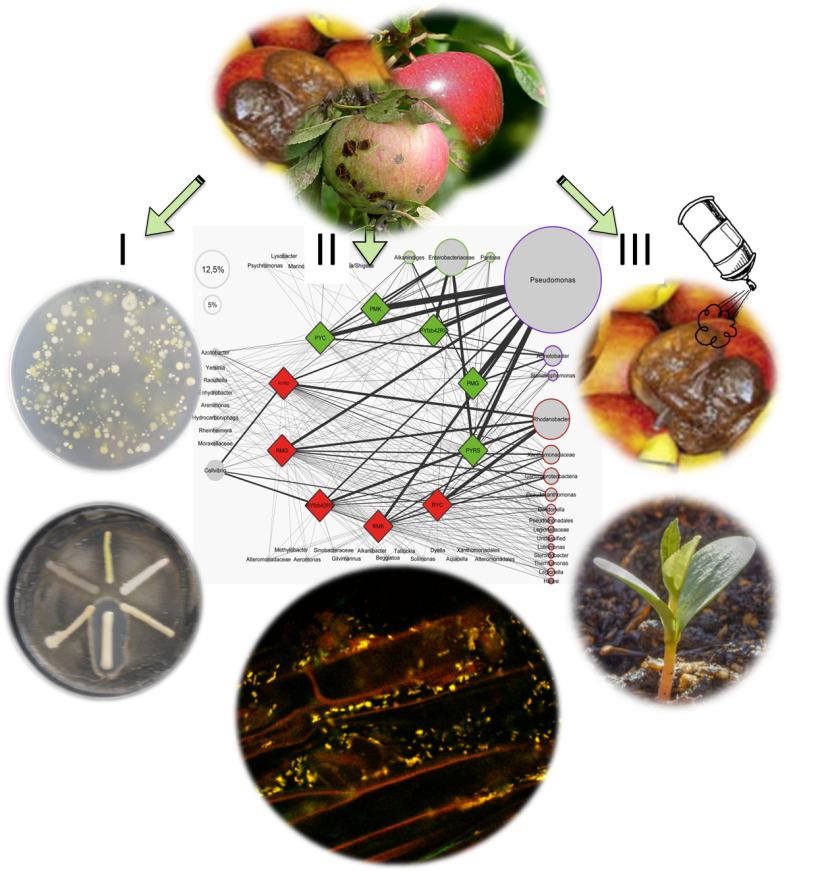
The experiments planned in the project are shown in Fig. 1. The experimental tasks will be carried out with selected students in small groups under the supervision of Master’s students and institute staff, with modules 1 and 6 contributing to the overall project results (see “Methods of Cooperation”).
Abb. 1.
Schema of the
scientific approach.
The microorganisms of healthy apples, as well as apple apples and apple rot are examined by three different methods:
(I)
Culture-Dependent Methods involve isolating the individual bacterial strains on an agar plate and then sequestering to select the plant disease antagonists.
(II)
The cultivation-independent methods capture the totality of microorganisms in the apple. The bacterial networks are visualized using bioinformatic methods (Fig. From Erlacher et al., 2014). Confocal laser scanning microscopy makes it possible to visualize the colonization patterns of natural apple-grown microorganisms.
(III)
Plant Model Tests will test the selected bacterial strains for their efficacy against plant diseases in planta .
Subsequently, environmentally friendly antimicrobials are tested against storage diseases.
Module 1
Ecological Excursions (years 1 and 2)
During the visits to SEKEM’s orchard, knowledge about sustainable agriculture is communicated through a variety of hands-on activities, thus bringing students problems and solutions. Plants are examined and leaf diseases are shown, as well as soil profiles are presented. The collected plant material is used to grow culturable microorganisms that live in the apples on agar plates and to detect the totality of microorganisms by sequencing techniques. This module is directly linked to module 6, in which the students document the sampling locations in detail, photograph and select the selected plant species and learn about them in this way.
Module 2
New microorganisms for plant protection (Year 1)
The bacteria isolated from apples in module 1 are selected by selected students in the laboratory of the TUG for their activity against the apple scab pathogen mold V. inaequalis, as well as the plant pathogen Verticillium longisporum (Rybakova et al., 2015, Fiss et al., 2000). All strains used in the studies are harmless to human health. Effective isolates are referred to as antagonists and can be identified, inter alia, by the formation of an inhibition zone in a dual culture assay. Such microorganisms that are capable of V. inaequalis and / or V. longisporum are closely examined microscopically and genetically (DNA sequencing).
The following questions can be answered:
1.) from which foods or plants were more antagonists obtained? and
2.) can differences in the number of antagonists in foods from organic and conventional cultivation be demonstrated?
Module 3 (Year 1)
Analysis of the Apple Microbiome
Organic apple trees (SEKEM ENERGY GmbH) as well as conventionally raised apples and those from private gardens of the pupils are being examined for disease.
Additional parameters are recorded, such as:
1.) Origin and mode of production: organic, conventional, private gardens or large-scale cultivation
2.) Age
3.) quality and pathogen infestation;
4.) Storage: freshly harvested or stored. These parameters can then be compared statistically with the data to identify the most important influence on the microbiome composition.
The work-up of the DNA samples comprises the following steps
1.) DNA isolation and
2.) Reproduction by PCR with specific primers for bacteria and archaea (515F / 806R, Caporaso et al., 2012),
3.) sequencing of the DNA,
4.) bioinformatory evaluation.
The scientific methods are described in Bragina et al. (2013) and Erlacher et al. (2014). The microorganisms in the apple are also visualized using confocal laser scanning microscopy. For this purpose, the samples are first fixed and then labeled with fluorescent DNA probes. In addition, by using LIVE / DEAD® BacLight Kit’s (ThermoFisher), a distinction can be made between live and dead bacteria, complementing the results of the microbiome study. As a result of the scientific investigation, bacterial libraries, network analyzes and an image archive with micro-organism micrographs of the apples are available.
The bioinformatics data are stored in international databases, where they can be used by researchers from all over the world and are available for further research projects.
Among other things, we will answer the following questions with the data obtained: 1.) which microorganism groups live in the apples we eat? How many live at the time of consumption?
2.) What is the difference between the microbiome of apples from organic and conventional crops, private gardens and large scale crops, and disease-prone and healthy trees?
3.) Which and how many of these microorganisms can be considered potentially pathogenic, or useful for humans or plants, and what do these differences mean for our health??
4.) Are there indicators for healthy apple trees?
5.) Which microorganisms are most common in the sick apple trees? These microorganisms are then bred in the follow-up projects (module 5 and summer internship).
Module 4 (Year 2)
New microorganisms against storage diseases
Microorganisms are isolated from the apples affected by rot pathogens. Together with the selected bacteria from the module 1 and the summer internships, they are tested for their ability to cause the Gloeosporium fungus pathogen Gloeosporium album and Gloeosporium perennans, to inhibit on an agar plate.
Module 5 (Year 2)
Testing of Microorganisms in Plant Models
The antagonists of plant pathogens isolated in modules 1 and 4 are additionally tested in plant models against the following pathogens as described in the corresponding literature:
1.) against apple rot pathogens in the apple model (Weiss et al 2006);
2.) against Venturia inaequalis in the apple leaf model (Yepes et al., 1993); and
3.) against Venturia inaequalis in the apple seedling model (Fiss et al, 2000). The microorganisms are additionally visualized using confocal laser scanning microscopy in vivo.
Summer Internships / Matura Theses
Evaluation of potential applications against plant and storage diseases. This module will be carried out by two students during a summer internship (Matura) and supervised by TUG staff and Roombiotic GmbH in collaboration with the storage company Meleco.
Organic apples are treated with a biological antimicrobial agent of natural origin, researched by Roombiotic (Figure 1).
After subsequent storage under commercial conditions, the treated apples are tested for the presence of storage diseases and compared to an untreated control group.
Subsequently, the infested apples are sorted out and used for the isolation of microorganisms associated with the disease.
The microorganisms are grown and their 16S rRNA gene fragment sequenced. The results of the bioinformatic evaluation (bacterial networks, module 2) are used to select the bacteria associated with the disease.
These selected strains are cultured and tested for sensitivity to the biological antimicrobial agent.
This provides a broader high throughput screening for beneficial microorganisms (plant pathogens antagonists), which in turn ensures a full quality control of the stored apples.
The data obtained will be used by the company Roombiotic and processed as part of one or more Matura thesis (s).
Module 6 (Years 1 and 2)
Artistic-Media Implementation
Following their research work within the project, students will participate in a professionally managed EcoArt art project that will make their findings accessible to the wider public and should meet with great media interest.
This enables a unique knowledge transfer that reaches all ages and populations.
The EcoArt exhibition will be held twice, at the end of every semester, and organized and organized by professional artist, sculptor and educator Timothy Mark.
The confocal microscope images,, which are created as part of the project, are enlarged and presented creatively as objects of art. In addition, a three-dimensional installation will be prepared with the help of students under the professional initiation and supervision of Mr. Mark, who will represent the colonization of the plant with the micro-organisms.
This sculpture is set up in a public square (for example the Botanical Garden in Graz) to serve as a stimulus for the population. This makes the invisible visible in a comprehensible form. In addition, a new approach to science will be created that will allow the younger audience to
It is planned that full grades, teachers, interested parents and siblings, as well as friends will actively participate in the exhibition preparation for the youth.
Above all, the EcoArt Day will serve to present scientific results creatively. In addition to PowerPoint, students learn how to use other media, such as self-produced images, 3D objects, films, music and the like.
Exciting hands-on experiments will be offered to the public during the EcoArt Day ( Stationslauf) in order to provide participants and guests with insights into everyday scientific life and to better understand the presented research topics. ,
This is organized by the company Biotenzz together with the TUG.